We leverage tools from machine learning and control theory to advance our understanding of biological systems. Control theoretic concepts are integrated both in the design of our optimization schemes and statistical machine learning models, as well as in the design of our in vitro and in vivo experiments. Current and future projects include
- Learning dynamics from biological time series
- Integrating multiple data modalities into a single model (including spatial modalities or the integration of prior knowledge from DNA or protein language models)
- Biological sequence model development (emphasis on long context sequence models, state space models)
- Designing microbial communities or bacteriotherapies (bugs-as-drugs) following control theoretic principles
- Gut-Brain axis
- Theoretical foundations at the intersection of machine learning and control
- Ultra-sensitive protein assay development and applications
Samples from our modeling research
To answer our biological questions surrounding the microbiome we build statistical models that propagate measurement and latent state uncertainty throughout the model. One example of this is our Robust and Scalable Model of Microbiome Dynamics (Figure 1). This model is a Bayesian nonparametric model of microbial dynamics based on what we term interaction modules, or learned clusters of latent variables with redundant interaction structure. This allows our model of dynamics to scale to hundreds of microbial taxa. Application of this model to “humanized” gnotobiotic mice can be found in our recent pre-print “Intrinsic instability of the dysbiotic microbiome revealed through dynamical systems inference at ecosystem-scale”. Another example of our modeling work is the method Chronostrain (pre-print). Chronostrain is a sequencing quality, host, and time aware strain tracking algorithm.

Sample from our experimental research
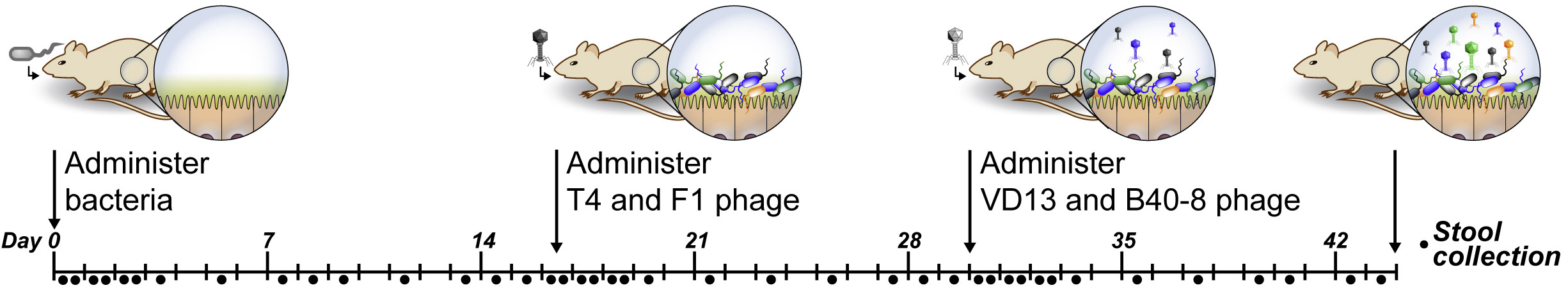
Our main experimental system for studying the microbiome is gnotobiotic mice. We are particularly interested in developing bacterio-therapies, and to develop those therapies we need to have an experimental system that allows us to probe the dynamics of complex microbial communities in vivo. We colonize germ-free mice with defined microbial communities as well as human fecal samples (creating “humanized” mice). To obtain rich dynamic profiles we perturb the microbial environment by changing the diet of the animals, presenting colonization challenges from other bacteria, delivering antibiotics, and by introducing bacteriophages. One of our experiments looking to understand the direct and indirect role of phage predation on the microbiome is shown in Figure 2 (accompanying paper).
Sample from our theoretical learning research
Gradient based optimization schemes are at the core of both adaptive control and optimization in ML (connections between the two are reported here). We leverage techniques used in adaptive control to improve the stability properties of momentum based gradient algorithms resulting in provably stable accelerated algorithms. Our algorithms are a log factor slower than Nesterov but with stability guarantees, see our Higher Order Tuner in Figure 3. Having a priori stability guarantees will be critical for certifying and deploying real-time and safety critical ML algorithms in medical applications.
